HiBC: The Human Intestinal Bacterial Collection
While Coscine assists researchers in their (meta)data management and collaboration, researchers may build upon this infrastructure. Here, we show how the Functional Microbiome Research Group at the University Hospital of RWTH Aachen share their data stored in Coscine.
The research group investigates the microbial communities associated with different animal hosts (e.g., mice, pig, human) by leveraging isolation and cultivation of bacterial isolates. Part of the characterisation of these isolates analyse their genomes obtained by sequencing their DNA. Metadata are collected throughout this data-generation process to provide relevant biological context and help assessing data quality. Therefore, data and metadata exploration and sharing are key steps in the investigation of the taxonomic identity and potential functions of these isolates.
On the Coscine side, Coscine's API is used to create a resource as well as uploading the data and metadata automatically. On the data sharing side, the researchers have made use of a R Shiny application that fetches the data via the S3 protocol in R. This accesses a read-only token for the S3 resource, allowing users from outside of the project to view and download the data.
(Meta)Data in Coscine
The researchers created a custom application profile to describe their data, namely the fasta (see profile in application profile generator). This allows for the full description of the data related to each isolate stored in Coscine.
All data is saved in a single S3 resource using this application profile. There are many ways to organize data in Coscine, but this approach has two notable benefits:
(1) It conserves resources, as the minimum resource size is 1 GB, while each bacterial isolate genome requires most of the time only up to 8 MB. (2) A single access token for the S3 resource (see below)
The researchers made use of the Coscine API along with the Python SDK to upload their (meta)data to the given resource. This accesses a metadata table, a result of the workflow creating the genomes, allowing for an automated metadata assignment.
To access the script, see information below.
Data Access
The S3 resource in Coscine allows the researchers to provide others data access using the read-only credentials. To facilitate this, they opted to avoid using S3 clients, instead using R and its package Shiny to build the HiBC web application. This provides a user-friendly interface:
Here, external researchers may view the HiBC's isolates, the associated cultivation conditions, explore the genome assemblies, and see further details for each isolate, as shown in the screenshots below. This draws on the metadata assigned to each dataset in Coscine using the above-mentioned application profile.
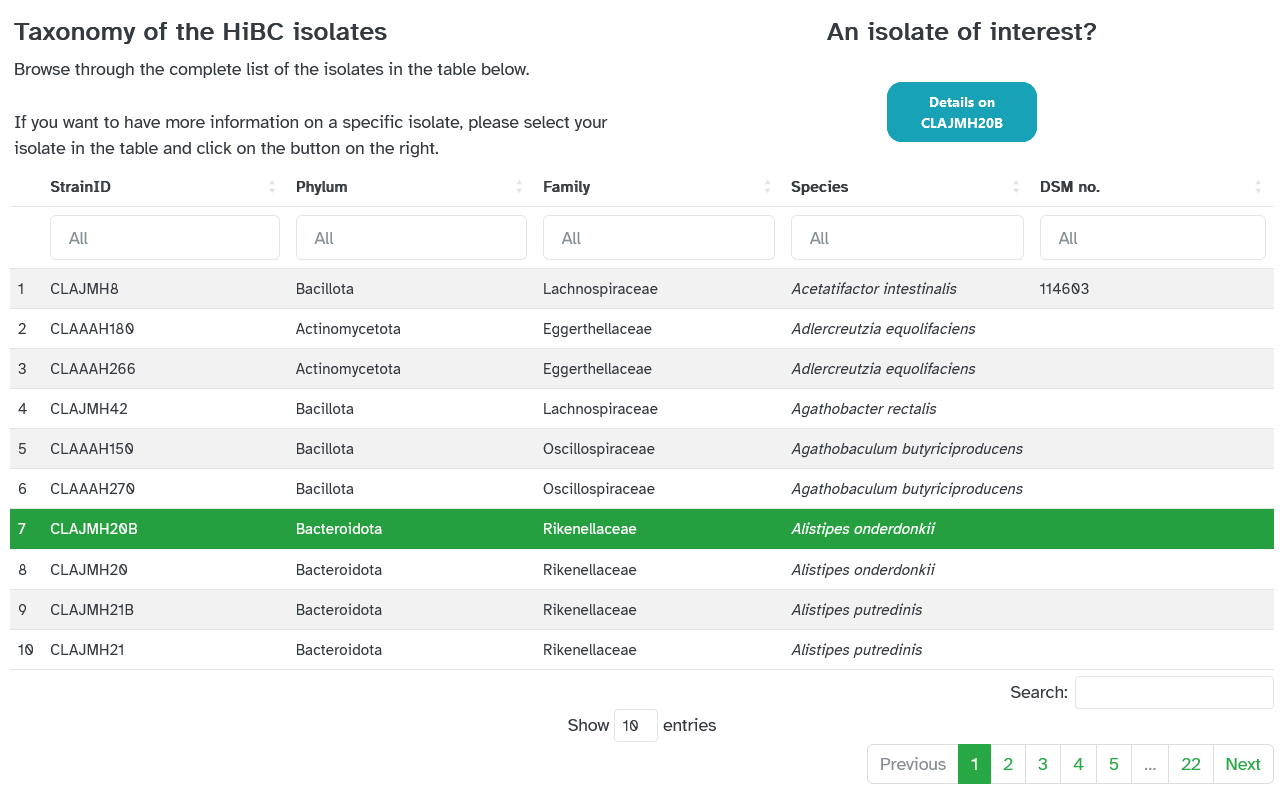
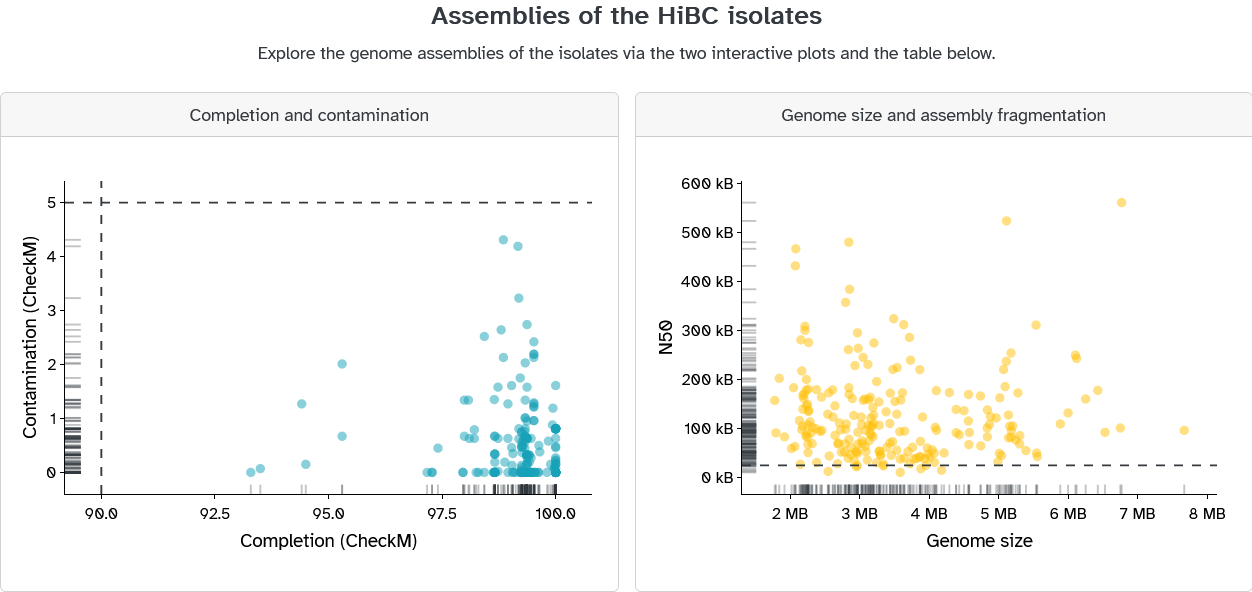

The associated FASTA file, stored in Coscine, may be downloaded for each isolate:

Sources and Further Information
Thomas C. A. Hitch, Johannes M. Masson, Charlie Pauvert, Johanna Bosch, Afrizal Afrizal, Nicole Treichel, Jonathan Hartman, Lukas C. Bossert and Thomas Clavel (n.d.). The Human Intestinal Bacterial Collection Website. Retrieved May 25, 2023 from https://hibc.otc.coscine.dev.
Code and information on the (meta)data transfer has been publicaly shared on GitLab.